Epidemiology of Enterococcus faecium isolates sampled from different sources in Romania using MLST technique and eBURST algorithm
DOI:
https://doi.org/10.55779/nsb15211546Keywords:
eBURST, Enterococcus faecium, epidemiology, MLSTAbstract
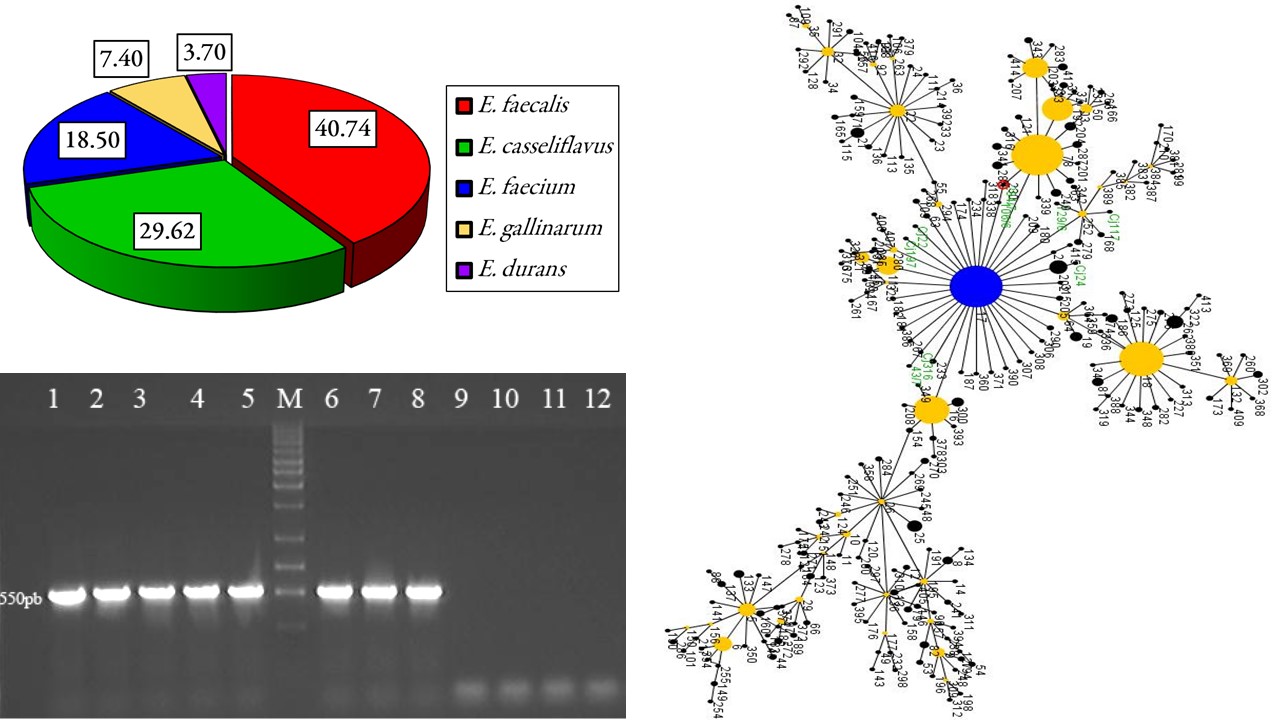
Enterococcus faecium is emerging as an important cause of multidrug resistance and hospital acquired infections, special attention being paid to the vancomycin resistant species. Therefore, the characterization of pathogenic strains/isolates plays an important role in the epidemiology of infectious diseases. The enterococcal rate was determined from wastewaters in Cluj-Napoca area. As presence of E. faecium was detected, a number of isolates from wastewater, birds and humans were epidemiologically analyzed according to the MLST website. Comparisons were performed against a collection of available isolates, with multiple origins, contained in the MLST database. Out of the Enterococcus isolates collected from wastewater, 11 were identified as E. faecalis (40.74%); 8 as E. casseliflavus (29.62%); 5 as E. faecium (18.50%); 2 as E. gallinarum (7.40%) and one isolate as E. durans. Based on the MLST data and using the eBURST algorithm, the isolates of E. faecium sampled from Romania were split in three groups: one group comprised isolates from human hosts and wastewater (Cj316, 106/6, Cj197, Cj22, 129/6, Cj117, Cj24, 284/7, and 43/7), while the second (G9, G10-2, G7, G3-2, and G9-1) and the third group (G8, G6, and 40/7) originated from bird hosts. The rest of the isolates were not joined in a particular group, assuming the lack of a phylogenetic bond between these isolates. The obtained data suggested the existence of at least two phylogenetic lines of E. faecium in Romania: a line that had mainly human host prevalence, while in the other line the animal hosts dominated.
Metrics
References
Adegoke AA, Madu CE, Reddy P, Stenström TA, Okoh AI (2022). Prevalence of vancomycin resistant Enterococcus in wastewater treatment plants and their recipients for reuse using PCR and MALDI-ToF MS. Frontiers in Environmental Science 9:797992. https://doi.org/10.3389/fenvs.2021.797992
Băcilă I, Jakab E, Popescu O (2007). Molecular characterization of Enterococcus faecium strains isolated from sewage using the multilocus sequence typing (MLST) technique. Annals NSBMB 16(3):69-77.
Blanch AR, Caplin JL, Iversen A, Kühn I, Manero A, Taylor HD, Vilanova X (2003). Comparison of enterococcal populations related to urban and hospital wastewater in various climatic and geographic European regions. Journal of Applied Microbiology 94(6):994-1002. https://doi.org/10.1046/j.1365-2672.2003.01919.x
Correa-Martínez CL, Jurke A, Schmitz J, Schaumburg F, Kampmeier S, Mellmann A (2022). Molecular epidemiology of vancomycin-resistant enterococci bloodstream infections in Germany: A population-based prospective longitudinal study. Microorganisms 10(1):130. https://doi.org/10.3390/microorganisms10010130
Dutka-Malen S, Even S, Courvalin P (1995). Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. Journal of Clinical Microbiology 35:24-27. https://doi.org/10.1128/jcm.33.1.24-27.1995
Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG (2004). eBURST: Infering patterns of evolutionary descends among clusters of related genotypes from multilocus sequence typing data. Journal of Bacteriology 186:1516-1530. https://doi.org/10.1128/JB.186.5.1518-1530.2004
Ferguson DM, Griffith JF, McGee CD, Weisberg SB, Hagedorn C (2013). Comparison of Enterococcus species diversity in marine water and wastewater using Enterolert and EPA Method 1600. Journal of Environmental and Public Health 2013:848049. https://doi.org/10.1155/2013/848049
Foulquié Moreno MR, Sarantinopoulos P, Tsakalidou E, de Vuyst L (2006). The role and application of enterococci in food and health. International Journal of Food Microbiology 106:1-24. https://doi.org/10.1016/j.ijfoodmicro.2005.06.026
Francisco AP, Bugalho M, Ramirez M, Carriço JA (2009). Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. https://doi.org/10.1186/1471-2105-10-152
Franz CMAP, Huch M, Abriouel H, Holzapfel W, Gálvez A (2011). Enterococci as probiotics and their implications in food safety. International Journal of Food Microbiology 151:125-140. https://doi.org/10.1016/j.ijfoodmicro.2011.08.014
Giraffa G (2014). Enterococcus. In: Batt CA, Tortorello ML (Eds). Encyclopedia of Food Microbiology (Second Edition). Academic Press, Amsterdam pp 674-679. https://doi.org/10.1016/B978-0-12-384730-0.00098-7.
Hall TA (1999). BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/95/NT. Nucleic Acids Symposium Series 41:95-98.
Heuer H, Smalla K (2007). Horizontal gene transfer between bacteria. Environmental Biosafety Research 6(1-2):3-13. https://doi.org/10.1051/ebr:2007034
Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, ... Willems RJL (2002). Multilocus sequence typing for Enterococcus faecium. Journal of Clinical Microbiology 40:1963-1971. https://doi.org/10.1128/JCM.40.6.1963-1971.2002
Karimi F, Samarghandi MR, Shokoohi R, Godini K, Arabestani MR (2016). Prevalence and removal efficiency of enterococcal species and vancomycin-resistant enterococci of a hospital wastewater treatment plant. Avicenna Journal of Environmental Health Engineering 3(2):e8623. https://doi.org/10.5812/ajehe.8623
Kim SH, Cho SY, Kim HM, Huh K, Kang C-I, Peck KR, Chung DR (2021). Sequence type 17 is a predictor of subsequent bacteremia in vancomycin-resistant Enterococcus faecium-colonized patients: a retrospective cohort study. Antimicrobial Resistance and Infection Control 10(1):108. https://doi.org/10.1186/s13756-021-00980-1
Klein G (2003). Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. International Journal of Food Microbiology 88:123-131. https://doi.org/10.1016/S0168-1605(03)00175-2
Kühn I, Iversen A, Burman LG, Olsson-Liljequist B, Franklin A, Finn M, ... Möllby R (2003). Comparison of enterococcal populations in animals, humans, and the environment – a European study. International Journal of Food Microbiology 88:133-145. https://doi.org/10.1016/S0168-1605(03)00176-4
Lebreton F, Willems RJL, Gilmore MS (2014). Enterococcus diversity, origins in nature, and gut colonization. In: Gilmore MS, Clewell DB, Ike Y, Shankar N (Eds). Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston pp 5-63.
Martínez-Carranza E, Barajas H, Alcaraz L-D, Servín-González L, Ponce-Soto G-Y, Soberón-Chávez G (2018). Variability of bacterial essential genes among closely related bacteria: the case of Escherichia coli. Frontiers in Microbiology 9:1059. https://doi.org/10.3389/fmicb.2018.01059
Moore DF, Guzman JA, McGee C (2008). Species distribution and antimicrobial resistance of enterococci isolated from surface and ocean water. Journal of Applied Microbiology 105(4):1017-1025. https://doi.org/10.1111/j.1365-2672.2008.03828.x
O’Driscoll T, Crank CW (2015). Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infection and Drug Resistance 8:217-230. https://doi.org/10.2147/IDR.S54125
Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J (2010). Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. The ISME Journal 4:377-387. https://doi.org/10.1038/ismej.2009.123
Pérez-Losada M, Browne EB, Madsen A, Wirth T, Viscidi RP, Crandall KA (2005). Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infection, Genetics and Evolution 6:97-112. https://doi.org/10.1016/j.meegid.2005.02.003
Ruoff KL, de la Maza L, Murtagh MJ, Spargo JD, Ferraro MJ (1990). Species identities of enterococci isolated from clinical specimens. Journal of Clinical Microbiology 28(3):435-437. https://doi.org/10.1128/jcm.28.3.435-437.1990
Shikov AE, Malovichko YV, Nizhnikov AA, Antonets KS (2022). Current methods for recombination detection in bacteria. International Journal of Molecular Sciences 23:6257. https://doi.org/10.3390/ijms23116257
Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Fridkin S, National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities (2013). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infection Control and Hospital Epidemiology 34(1):1-14. https://doi.org/10.1086/668770
Sinton LW, Donnison AM (1994). Characterisation of faecal streptococci from some New Zealand effluents and receiving waters. New Zealand Journal of Marine and Freshwater Research 28(2):145-158. https://doi.org/10.1080/00288330.1994.9516603
Spratt BG, Hanage WP, Li B, Aanensen DM, Feil EJ (2004). Displaying the relatedness among isolates of bacterial species – the eBurst approach. FEMS Microbiology Letters 241:129-134. https://doi.org/10.1016/j.femsle.2004.11.015
Stefani S, Agodi A (2000). Molecular epidemiology of antibiotic resistance. International Journal of Antimicrobial Agents 13:143-153. https://doi.org/10.1016/S0924-8579(99)00103-X
Stern CS, Carvalho M da G, Teixeira LM (1994). Characterization of enterococci isolated from human and nonhuman sources in Brazil. Diagnostic Microbiology and Infectious Disease 20(2):61-67. https://doi.org/10.1016/0732-8893(94)90093-0
Tavanti A, Davidson AD, Johnson EM, Maiden MCJ, Shaw DJ, Gow NAR, Odds FC (2005). Multilocus sequence typing for differentiation of strain of Candida tropicalis. Journal of Clinical Microbiology 43:5993-5600. https://doi.org/10.1128/JCM.43.11.5593-5600.2005
Toc DA, Butiuc-Keul AL, Iordache D, Botan A, Mihaila RM, Costache CA, ... Junie LM (2022). Descriptive analysis of circulating antimicrobial resistance genes in vancomycin-resistant Enterococcus (VRE) during the COVID-19 Pandemic. Biomedicines 10:1122. https://doi.org/10.3390/biomedicines10051122
Willems RJL, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, .... Bonten MJM (2005). Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerging Infectious Diseases 11(6):821-8. https://doi.org/10.3201/eid1106.041204
Zhong Z, Zhang W, Song Y, Liu W, Xu H, Xi X, ... Sun Z (2017). Comparative genomic analysis of the genus Enterococcus. Microbiological Research 196:95-105. https://doi.org/10.1016/j.micres.2016.12.009
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Ioan BĂCILĂ, Endre JAKAB, Dana ŞUTEU, Octavian POPESCU

This work is licensed under a Creative Commons Attribution 4.0 International License.
Papers published in Notulae Scientia Biologicae are Open-Access, distributed under the terms and conditions of the Creative Commons Attribution License.
© Articles by the authors; licensee SMTCT, Cluj-Napoca, Romania. The journal allows the author(s) to hold the copyright/to retain publishing rights without restriction.
License:
Open Access Journal - the journal offers free, immediate, and unrestricted access to peer-reviewed research and scholarly work, due SMTCT supports to increase the visibility, accessibility and reputation of the researchers, regardless of geography and their budgets. Users are allowed to read, download, copy, distribute, print, search, or link to the full texts of the articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author.













.png)















