Introgression progress for phenotypic traits and parent-progeny diversity at advanced segregation population from Oryza barthii and O. glaberrima/O. sativa crosses
Keywords:
advanced population, gene introgression, inter-generation diversity, Oryza barthii, parent-progeny correlation, segregationAbstract
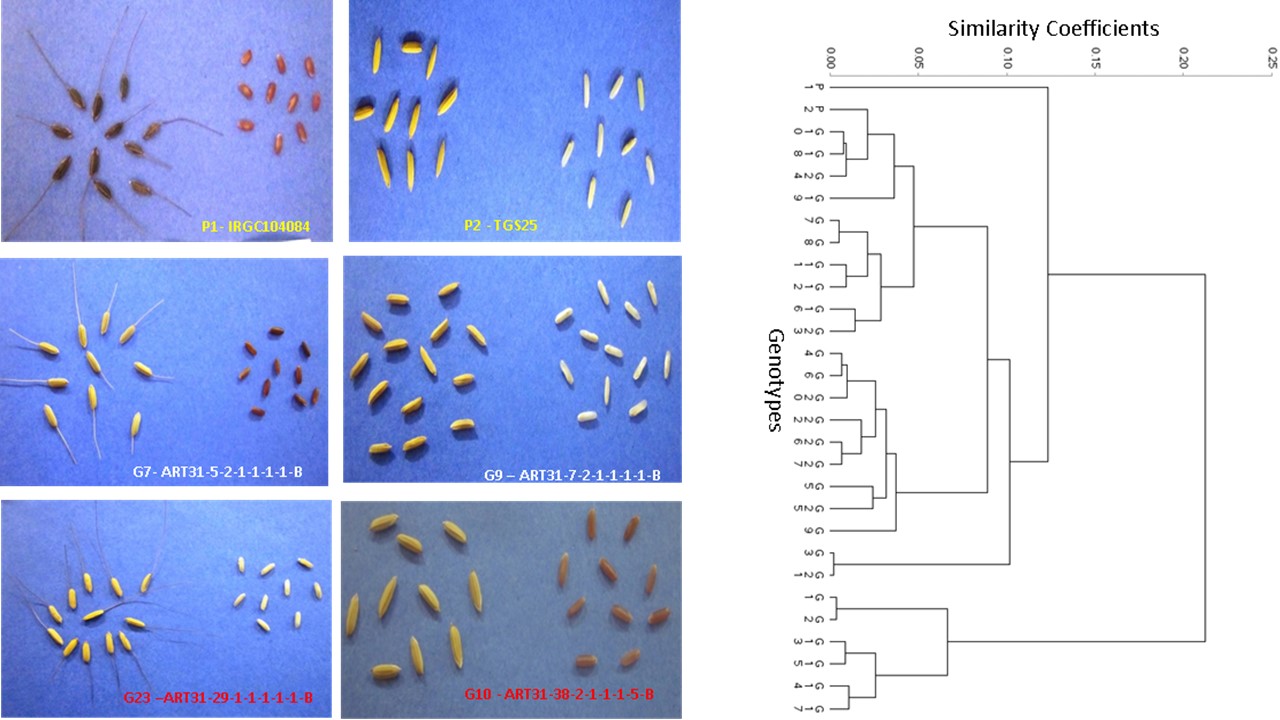
Oryza barthii has candidature for some significant economic traits but its utilization in rice breeding is rare. This study traced introgression of heritable traits in the offspring of O. barthii with an Africa-Asian progenitor to F8 and assessed diversity between the parents and the F8 population. Significant (P<0.05) genotypic variation existed for some traits. Grains per panicle and days to 50% flowering had respective least (3.34%) and highest (96.32%) broad sense heritability. Genotypic Coefficient of Variation (GCV) was lower than Phenotypic Coefficient of Variation (PCV) in all traits. Grains per panicle and tiller number had respective least (5.28% and 8.05%) and highest (90.8% and 98.1%) GCV and PCV. Progenies significantly differ in panicles and grains sizes, shapes, colours, presence or absence of awns. Five principal components explained 80.1% of the total variance. Plant height at maturity was the only trait with significant (p ≤ 0.01) correlation and regression between F6 and F7. Progenies resemblance to P1 retrogressively declined while offspring similarity to P2 progressively increased from F6 to F8. The present diversity study discovered three heterotic groups: the O. barthii (11%), O. sativa (67%) and the intermediate group (22%). This research has added to rice genetic resources, making investigation of the nutritional status of the different progenies interesting research for further studies.
Metrics
References
Aananthi N (2018). Inter generation trait association and regression analysis in f2 and f3 generations of rice. International Journal of Current Microbiology and Applied Sciences 7:3651-3662. https://doi.org/10.20546/ijcmas.2018.708.370.
Africa Rice Center (AfricaRice) (2012). A new rice research for development strategy for Africa. Africa Rice Center annual report 2011 Cotonou, Benin.
Falconer DS, Mackay TFC (1996). Introduction to quantitative genetics. Longman scientific & technical ltd, Essex.
Govintharaj P, Tannidi S, Swaminathan M, Sabariappan R (2017). Effectiveness of selection, parent- offspring correlation and regression in bacterial blight resistance genes introgressed rice segregating population. Ciência Rural 47:09-14. http://dx.doi.org/10.1590/0103-8478cr20160987.
Gujja B, Thiyagarajan TM (2009). New hope for Indian food security? The system of rice intensification. The Gatekeeper Series 143:1-18.
Ikeda R (2004). For the development of sustainable rice cultivation in Africa. JIRCAS. Newsletter 38:22-28.
IRRI (2005). Research paper series. IRRI. Los Baños, Philippines.
Janwan M, Sreewongchai T, Sripichitt P (2013). Rice breeding for high yield by advanced single seed descent method of selection. Journal of Plant Science 8:24-30. https://doi.org/10.3923/jps.2013.24.30.
Jin X, Chen Y, Liu P, Li C, Cai XX, Rong J, Lu BR (2018). Introgression from cultivated rice alters genetic structures of wild relative populations: implications for in situ conservation. AoB Plants 10:plx055. https://doi.org/10.1093/aobpla/plx055.
Kahani F, Hittalmani S (2016). Identification of f2 and f3 segregants of fifteen rice crosses suitable for cultivation under aerobic situation. Sabrao Journal of Breeding and Genetics 48:219-229.
Kanbar A, Kondo K, Shashidhar HE (2011). Comparative efficiency of pedigree, modified bulk and single seed descent breeding methods of selection for developing high-yielding lines in rice (Oryza sativa L.) under aerobic condition. Electronic Journal of Plant Breeding 2:184-193.
Khush GS (1997). Origin, dispersal, cultivation and variation of rice. Plant Molecular Biology 35:25-34.
Lin Z, Qin P, Zhang X, Fu C, Deng H, Fu X, … Deng XW (2020). Divergent selection and genetic introgression shape the genome landscape of heterosis in hybrid rice. Proceedings of the National Academy of Sciences 117:4623-4631. https://doi.org/10.1073/pnas.1919086117
Li KS (1970). The origin of cultivated plants in Southeast Asia. Economic Botany 24:3-19. https://doi.org/10.1007/BF02860628
Liu D, Wang J, Wang X, Yang XJ, Sun J, Chen W (2015). Genetic diversity and elite gene introgression reveal the japonica rice breeding in northern China. Journal of Integrative Agriculture 14:811-822. https://doi.org/10.1016/S2095-3119(15)61050-4
Li ZM, Zheng XM, Ge S (2011). Genetic diversity and domestication history of African rice (Oryza glaberrima) as inferred from multiple gene sequences. Theoretical and Applied Genetics 123:21-31. Https://doi.org/10.1007/s00122-011-1563-2
Linares OF (2002). African rice (Oryza glaberrima); history and future potential. Proceedings of the National Academy of Sciences of the United States of America 99:16360-16365. https://doi.org/10.1073/pnas.252604599.
MacLean JL, Dawe DC, Hardy B, Hettel GP (2002). Rice almanac. Los Banos Philippines. https://doi.org/10.1093/aob/mcg189
Maricel AB (2010). Genetic analysis of agronomic traits, yield components and grain quality in Oryza barthii derivatives. CIAT Annual Report.
Matsuo T (1997). Science of the rice plant. Food and agriculture policy research center.
Mohammadi SA, Prasanna B (2003). Analysis of genetic diversity in crop plants—salient statistical tools and considerations. Crop Science 43:1235-1248. https://doi.org/10.1186/s12864-017-3922-0
Mohanty S (2009). Why global rice production is plunging. CommodityOnline, available at: http://www.commodityonline.com/news/why-global-rice-production-is-plunging-13800-2-1.html.
National Research Council (1996). Lost crops of Africa: Grains. National Academic Press, Washington, DC, Vol. 1, pp 17.
Ogunbayo SA, Sie M, Ojo DK, Sanni KA, Akinwale MG, Toulou B, Shittu A, Dehen EO, Popoola AR, Daniel IO, Gregorio GB (2014). Genetic variation and heritability of yield and related traits in promising rice genotypes (Oryza sativa L.). Journal of Plant Breeding and Crop Science 6:153-159. https://doi.org/10.5897/JPBCS2014.0457
Oka HI (1988). Origin of cultivated rice. Elsevier science publishers, pp 129.
Prajapati M, Singh CM, Suresh BG, Lavanya GR, Jadhav P (2011). Genetic parameters for grain yield and its component characters in rice. Electronic Journal of Plant Breeding 2(2):235-238.
Prasad B, Patwari AK, Biswas PS (2001). Genetic Variability and selection criteria in fine grain rice (Oryza sativa). Pakistan Journal of Biological Sciences 4(10):1188-1190. https://doi.org/10.3923/pjbs.2001.1188.1190
Sampath S (1973). Origins of cultivated rice. Indian Journal of Genetics and Plant Breeding 33:157-161.
Sarla N, Swamy BPM (2005). Oryza glaberrima: a source for the improvement of Oryza sativa. Current Science 89:955-963.
SAS (2011). SAS procedures guide. 9.4 edition. SAS Institute Inc., Cary, NC. USA.
Scott RA, Milliken GA (1993). A SAS program for analyzing augmented randomized complete block designs. Crop Science 33:865-867. https://dx.doi.org/10.2135/cropsci1993.0011183X003300040046x
Standard Evaluation System for Rice (SES) (2002) International Rice Research Institute, (IRRI). Los Baños, Philippines.
STAR (2014). STAR Software version 2.0.1. Biometrics and Breeding Informatics. BPGB Division, International Rice Research Institute, Los Banos, Laguna.
Sürek H, Korkut KZ, Bilgin O (1998). Correlation and path analysis for yield and yield components in rice in an 8-parent diallel set of crosses. Oryza 35:15-18.
Vanniarajan AC, Ramalingam J (2011). Parent Progeny regression analysis in f2 and f3 generations of rice. Electronic Journal of Plant Breeding 2:520-522.
Wolf JB, Wade MJ (2009). What are maternal effects and what are they not? Philosophical Transactions of the Royal Society B: Biological Sciences 364(1520):1107-1115. https://doi.org/10.1098/rstb.2008.0238
Yadav P, Rangare NR., Anurag PJ, Chaurasia AK (2007). Quantitative analysis of rice (Oryza sativa L.) in Allahabad agro-climatic zone. Journal of Rice Research 3:16-18.
Yan W, Kang MS (2003). GGE Biplot Analysis: A graphical tool for breeders, geneticists, and agronomists. CRC press, Boca Raton, FL.
Downloads
Published
How to Cite
Issue
Section
License
Papers published in Notulae Scientia Biologicae are Open-Access, distributed under the terms and conditions of the Creative Commons Attribution License.
© Articles by the authors; licensee SMTCT, Cluj-Napoca, Romania. The journal allows the author(s) to hold the copyright/to retain publishing rights without restriction.
License:
Open Access Journal - the journal offers free, immediate, and unrestricted access to peer-reviewed research and scholarly work, due SMTCT supports to increase the visibility, accessibility and reputation of the researchers, regardless of geography and their budgets. Users are allowed to read, download, copy, distribute, print, search, or link to the full texts of the articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author.









.png)















