In silico testing of C9H12ClNO2 and C6H5Cl2NO as derivatives of acetaminophen using molecular docking method
DOI:
https://doi.org/10.55779/nsb16111632Keywords:
active region, computational approach, fever reduction, functional group alteration, ligand-receptor recognition, molecular docking, paracetamol, pharmacological characteristics, therapeutic efficacyAbstract
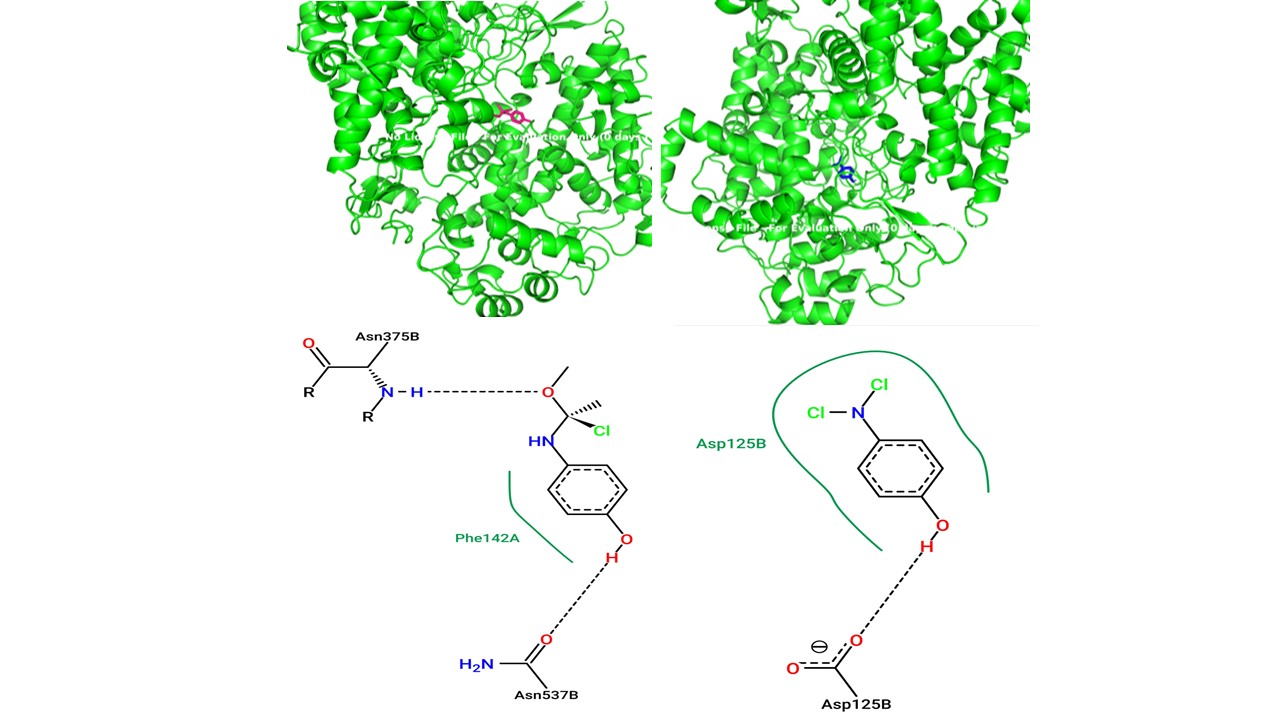
Paracetamol, a commonly used analgesic and antipyretic medication, is well-known for its ability to relieve pain and reduce temperature. However, there is a constant push to improve its therapeutic efficacy, especially towards increasing its oral bioavailability. The increase in bioavailability will lead to a better reception of the drugs by the body. This research aims to provide valuable insights into the molecular mechanisms underlying paracetamol’s mode of action and propose novel strategies for enhancing its therapeutic effectiveness. We investigated the notion of functional group alteration by molecular docking as a strategy to increase the efficacy of paracetamol in this work. Using modern computational approaches, it could be conducted through the examination of the structural characteristics and active regions of paracetamol and its target receptors. Additionally, molecular docking simulations were used to examine the binding interactions between paracetamol and its target receptors, offering insights into the essential functional groups required for ligand-receptor recognition. Tests of several molecular docking techniques and scoring functions allowed the researchers to find potential alterations that might improve its pharmacological characteristics. By integrating structural analysis, molecular docking studies, and computational screening, the uncovering of promising modifications that can significantly improve paracetamol’s efficacy was expected. Ultimately, this work may lead to the development of next-generation analgesics with superior pharmacological profiles, providing enhanced pain relief and fever reduction for patients.
Metrics
References
Aini NS, Ansori ANM, Kharisma VD, Murtadlo AAA, Tamam MB, Sucipto TH, ... Zainul R (2023a). An in silico study: Phytochemical compounds screening of garcinia atroviridis griff. ex t. anders as anti-DENV. Journal of Pure and Applied Microbiology 17(4):2467-2478. https://doi.org/10.22207/jpam.17.4.45
Aini NS, Kharisma VD, Ansori A, Murtadlo AAA, Tamam MB, Turista DDR, ... Zainul R (2023b). Bioactive compounds screening of Rafflesia sp. and Sapria sp. (Family: Rafflesiaceae) as anti-SARS-CoV-2 via tetra inhibitors: An in silico research. Journal of Pharmacy and Pharmacognosy Research 11(4):611-624. https://doi.org/10.56499/jppres23.1620_11.4.611
Atkinson HC, Stanescu I, Frampton C, Salem II, Beasley CP, Robson R (2015). Pharmacokinetics and bioavailability of a fixed-dose combination of ibuprofen and paracetamol after intravenous and oral administration. Clinical Drug Investigation 35:625-632. https://doi.org/10.1007/s40261-015-0320-8
Ayoub SS (2021). Paracetamol (acetaminophen): A familiar drug with an unexplained mechanism of action. Temperature 8(4):351-371. https://doi.org/10.1080/23328940.2021.1886392
Bhattacharya D, Cheng J (2013). 3Drefine: Consistent protein structure refinement by optimizing hydrogen bonding network and atomic‐level energy minimization. Proteins: Structure, Function, and Bioinformatics 81(1):119-131. https://doi.org/10.1002/prot.24167
Chung CW, Dean AW, Woolven JM, Bamborough P (2012). Fragment-based discovery of bromodomain inhibitors part 1: inhibitor binding modes and implications for lead discovery. Journal of Medicinal Chemistry 55(2):576-586. https://doi.org/10.1021/jm201320w
Dallakyan S, Olson AJ (2015). Small-molecule library screening by docking with PyRx. Chemical Biology: Methods and Protocols 243-250. https://doi.org/10.1007/978-1-4939-2269-7_19
Daneshgar P, Moosavi-Movahed AA, Norouzi P, Ganjali MR, Madadkar-Sobhani A, Saboury AA (2009). Molecular interaction of human serum albumin with paracetamol: spectroscopic and molecular modeling studies. International Journal of Biological Macromolecules 45(2):129-134. https://doi.org/10.1016/j.ijbiomac.2009.04.011
Dayer MR (2020). Old drugs for newly emerging viral disease, COVID-19: Bioinformatic Prospective. arXiv preprint arXiv:2003.04524. https://doi.org/10.48550/arXiv.2003.04524
El-Shaheny R, Fuchigami T, Yoshida S, Radwan MO, Nakayama M (2019) Complementary HPLC, in silico toxicity, and molecular docking studies for investigation of the potential influences of gastric acidity and nitrite content on paracetamol safety. Microchemical Journal 150:104107. https://doi.org/10.1016/j.microc.2019.104107
Fährrolfes R, Bietz S, Flachsenberg F, Meyder A, Nittinger E, Otto T, ... Rarey, M (2017). Proteins Plus: a web portal for structure analysis of macromolecules. Nucleic Acids Research 45(W1):W337-W343. https://doi.org/10.1093/nar/gkx333
Ferreira GE, Dmitritchenko A, McLachlan AJ, Day RO, Saragiotto B, Lin C, ... Maher, C. G (2021). The efficacy and safety of paracetamol for pain relief: an overview of systematic reviews. The Medical Journal of Australia 214(7):324-331. https://doi.org/10.5694/mja2.50992
Ghlichloo I, Gerriets V (2019). Nonsteroidal anti-inflammatory drugs (NSAIDs). Europe PMC; StatPearls Publishings. https://europepmc.org/article/nbk/nbk547742
Grogan S, Preuss CV (2021). Pharmacokinetics. Nih.gov; StatPearls Publishing. https://www.ncbi.nlm.nih.gov/books/NBK557744/
Grosser T, Theken KN, FitzGerald GA (2017). Cyclooxygenase inhibition: pain, inflammation, and the cardiovascular system. Clinical Pharmacology and Therapeutics 102(4):611-622. https://doi.org/10.1002/cpt.794
Hannibal CG, Dehlendorff C, Kjaer SK (2018). Use of paracetamol, low-dose aspirin, or non-aspirin non-steroidal anti-inflammatory drugs and risk of ovarian borderline tumors in Denmark. Gynecologic Oncology 151(3):513-518. https://doi.org/10.1016/j.ygyno.2018.09.022
Jain AS, Sushma P, Dharmashekar C, Beelagi MS, Prasad SK, Shivamallu C, ... Prasad KS (2021). In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2. Saudi Journal of Biological Sciences 28(1):1040-1051. https://doi.org/10.1016/j.sjbs.2020.11.049
Kharisma VD, Ansori ANM, Antonius Y, Rosadi I, Affan A, Jakhmola V, ... Purnobasuki H. (2023). Garcinoxanthones from Garcinia mangostana L. against sars-cov-2 infection and cytokine storm pathway inhibition: A viroinformatics study. Journal of Pharmacy and Pharmacognosy Research 11(5):743-756. https://doi.org/10.56499/jppres23.1650_11.5.743
Lobo, S (2019). Is there enough focus on lipophilicity in drug discovery? Expert Opinion on Drug Discovery 15(3):261-263. https://doi.org/10.1080/17460441.2020.1691995
Lonsdale R, Ward RA (2018). Structure-based design of targeted covalent inhibitors. Chemical Society Reviews 47(11):3816-3830. https://doi.org/10.1039/C7CS00220C
Manikandan P, Nagini S (2018). Cytochrome P450 Structure, Function and Clinical Significance: A Review. Current Drug Targets 19(1). https://doi.org/10.2174/1389450118666170125144557
McCrae JC, Morrison EE, MacIntyre IM, Dear JW, Webb DJ (2018). Long‐term adverse effects of paracetamol–a review. British Journal of Clinical Pharmacology 84(10):2218-2230. https://doi.org/10.1111/bcp.13656
Mooers BH (2016). Simplifying and enhancing the use of PyMOL with horizontal scripts. Protein Science 25(10):1873-1882. https://doi.org/10.1002/pro.2996
Moriarty C, Carroll W (2016). Paracetamol: pharmacology, prescribing and controversies. Archives of Disease in Childhood-Education and Practice 101(6):331-334. http://dx.doi.org/10.1136/archdischild-2014-307287
Oloyede OO, Alabi ZO, Akinyemi AO, Oyelere SF, Oluseye AB, Owoyemi BCD (2023). Comparative evaluation of acetaminophen form (I) in commercialized paracetamol brands. Scientific African 19:e01537. https://doi.org/10.1016/j.sciaf.2022.e01537
Pacifici GM, Allegaert K (2015). Clinical pharmacology of paracetamol in neonates: a review. Current Therapeutic Research 77:24-30. https://doi.org/10.1016/j.curtheres.2014.12.001
Padmi H, Kharisma VD, Ansori ANM, Sibero MT, Widyananda MH, Ullah ME, ... Zainul R (2022). Macroalgae bioactive compounds for the potential antiviral of SARS-CoV-2: An in silico study. Journal of Pure and Applied Microbiology 16(2):1018-1027. https://doi.org/10.22207/jpam.16.2.26
Price G, Patel DA (2020). Drug Bioavailability. PubMed; StatPearls Publishing. https://www.ncbi.nlm.nih.gov/books/NBK557852/
Qureshi MA, Khan TJ, Qureshi MS (2011). Molecular Docking helps in understanding the action of Paracetamol (acetaminophen): an approach towards finding a better COX2 inhibitor. RMJ 36(3):238-241. https://www.rmj.org.pk/fulltext/27-1298307061.pdf
Ralapanawa U, Jayawickreme KP, Ekanayake EMM, Dissanayake AMS (2016). A study on paracetamol cardiotoxicity. BMC Pharmacology and Toxicology 17(1):1-8. https://doi.org/10.1186/s40360-016-0073-x
Sanches BMA, Ferreira EI (2019). Is prodrug design an approach to increase water solubility? International Journal of Pharmaceutics 568:118498. https://doi.org/10.1016/j.ijpharm.2019.118498
Sharma CV, Mehta V (2014). Paracetamol: mechanisms and updates. Continuing Education in Anaesthesia, Critical Care and Pain 14(4):153-158. https://doi.org/10.1093/bjaceaccp/mkt049
Schrödinger LLC (2010). The PyMOL molecular graphics system. version 2.5.0
Skoraczyński G, Kitlas M, Miasojedow B, Gambin A (2023). Critical assessment of synthetic accessibility scores in computer-assisted synthesis planning. Journal of Cheminformatics 15(1). https://doi.org/10.1186/s13321-023-00678-z
Supe S, Takudage P (2020). Methods for evaluating penetration of drug into the skin: A review. Skin Research and Technology 27(3). https://doi.org/10.1111/srt.12968
Tzankova V, Aluani D, Kondeva-Burdina M, Yordanov Y, Odzhakov F, Apostolov A, Yoncheva K (2017). Hepatoprotective and antioxidant activity of quercetin loaded chitosan/alginate particles in vitro and in vivo in a model of paracetamol-induced toxicity. Biomedicine and Pharmacotherapy 92:569-579. https://doi.org/10.1016/j.biopha.2017.05.008
Uzzaman M, Shawon J, Siddique ZA (2019). Molecular docking, dynamics simulation and ADMET prediction of Acetaminophen and its modified derivatives based on quantum calculations. SN Applied Sciences 1:1-10. https://doi.org/10.1007/s42452-019-1442-z
Van Rensburg R, Reuter H (2019). An overview of analgesics: NSAIDs, paracetamol, and topical analgesics Part 1. South African Family Practice. https://doi.org/10.1080/20786190.2019.1610228
Villa-Zapata L, Gomez-Lumbreras A, Horn JR, Tan M, Boyce RD, Malone DC (2022). A Disproportionality Analysis of Drug–Drug Interactions of Tizanidine and CYP1A2 Inhibitors from the FDA Adverse Event Reporting System (FAERS). Drug Safety 45(8):863-871. https://doi.org/10.1007/s40264-022-01200-4
Wahyuni DK, Wacharasindhu S, Bankeeree W, Punnapayak H, Purnobasuki H, Junairiah, ... Prasongsuk S (2022). Molecular simulation of compounds from n-hexane fraction of Sonchus Arvensis L. leaves as SARS-CoV-2 antiviral through inhibitor activity targeting strategic viral protein. Journal of Pharmacy and Pharmacognosy Research 10(6):1126-1138. https://doi.org/10.56499/jppres22.1489_10.6.1126
Wakefield AE, Kozakov D, Vajda S (2022). Mapping the binding sites of challenging drug targets. Current Opinion in Structural Biology 75:102396. https://doi.org/10.1016/j.sbi.2022.102396
Wang Y, Lin W, Wu N, He X, Wang J, Feng Z, Xie XQ (2018). An insight into paracetamol and its metabolites using molecular docking and molecular dynamics simulation. Journal of Molecular Modeling 24:1-13. https://doi.org/10.1007/s00894-018-3790-9
Yang J, Anishchenko I, Park H, Peng Z, Ovchinnikov S, Baker D (2020). Improved protein structure prediction using predicted interresidue orientations. Proceedings of the National Academy of Sciences 117(3):1496-1503. https://doi.org/10.1073/pnas.1914677117
Yuan S, Chan HCS, Hu Z (2017). Using PyMOL as a platform for computational drug design. Wiley Interdisciplinary Reviews: Computational Molecular Science 7(2):e1298. https://doi.org/10.1002/wcms.1298
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Juan LORELL, Kathy IVANA, Joselyn P.W. TANOTO, Michael MICHAEL, Sarah JESCIKA, Nicolaas R.P. GAUTAMA, Olivia TJOA, Keisha ALINA, Arli A. PARIKESIT, Fandi SUTANTO

This work is licensed under a Creative Commons Attribution 4.0 International License.
Papers published in Notulae Scientia Biologicae are Open-Access, distributed under the terms and conditions of the Creative Commons Attribution License.
© Articles by the authors; licensee SMTCT, Cluj-Napoca, Romania. The journal allows the author(s) to hold the copyright/to retain publishing rights without restriction.
License:
Open Access Journal - the journal offers free, immediate, and unrestricted access to peer-reviewed research and scholarly work, due SMTCT supports to increase the visibility, accessibility and reputation of the researchers, regardless of geography and their budgets. Users are allowed to read, download, copy, distribute, print, search, or link to the full texts of the articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author.













.png)















