Phylogenetic assessment and in silico characterization of cytochrome b protein of three alpheid shrimps
DOI:
https://doi.org/10.15835/nsb14111202Keywords:
cyt b, phylogeny, physicochemical characters, Ramachandran plot, snapping shrimpAbstract
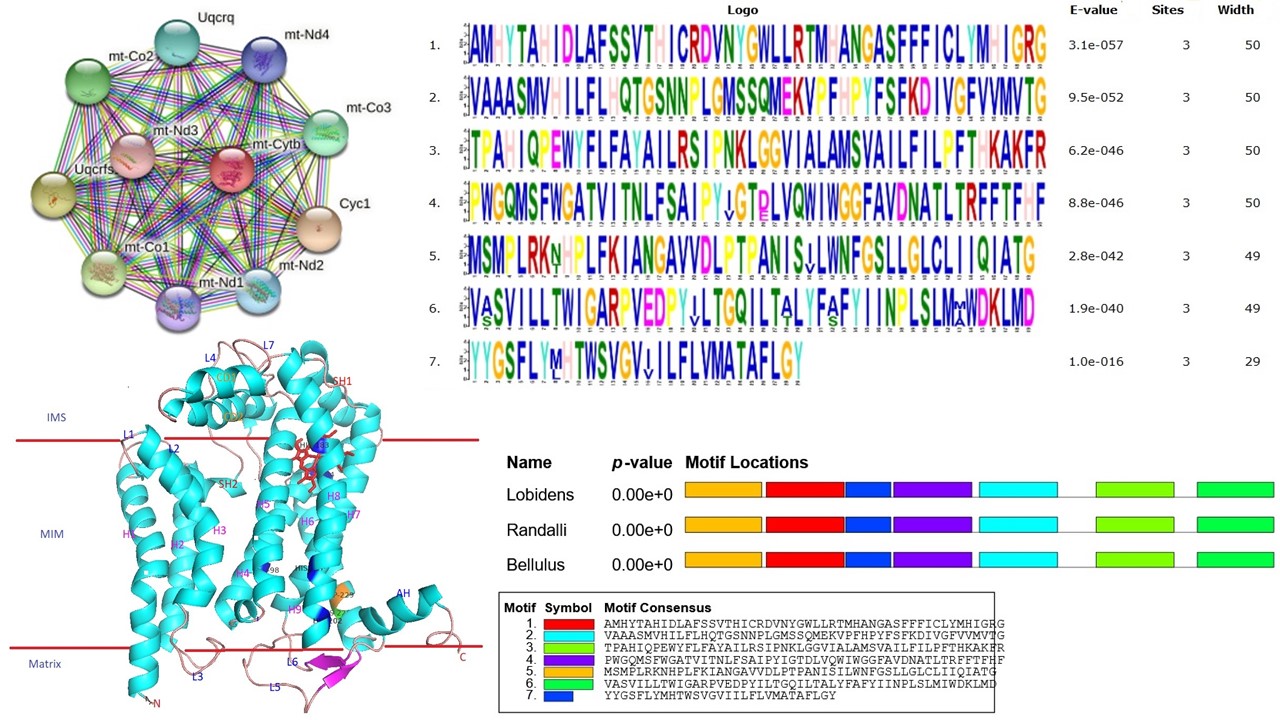
Cytochrome b (cyt b) is one of the cytochrome proteins involved in electron transport in the respiratory chain of mitochondria. Cyt b is the only gene among the cytochrome complex coded by mitochondrial DNA. It is the most widely used gene for phylogenetic assessment and inter species variation studies. Here, the amino acid sequence of cyt b in three snapping shrimps such as, Alpheus lobidens, A. randali, A. bellulus was analysed and the results showed higher similarity in A. lobidens and A. randali as reflected in the phylogenetic tree. This study describes the applications of bioinformatics tools to predict the physico-chemical characters of cyt b protein. This protein was composed of least percentage of Cys (0.8%) and highest percentage of Leu (13.8%). The maximum molecular weight (MW) was predicted as 42.62 KDa in A. randali. The theoretical pI value was ranged from 8.35 to 8.36 and confirmed that cyt b was alkali in nature. The instability index value was in the range of 42.29 to 46.94 which showed the protein was more stable. The secondary structure of this protein was primarily composed of α-helixes and random coil, revealing the stable structure. The comparative modelling was performed by Swiss model where the 3-D crystal structure of bovine cyt bc1 (6haw1.c) was used as template. Ramachandran plot analysis showed that most of the amino acids (>92%) falling on the favoured region. Seven conserved motifs were identified by MEME analysis. The modelled 3-D structure of this protein was validated by PROCHECK and QMEAN. The transmembrane protein topology and helix probability curve was predicted by TMHMM server. Protein-protein interactions was analysed by STRING tool and found the network of cyt b with related proteins. The results of this study may provide valuable insights into fundamental characteristics of cyt b in Alpheid shrimps.
Metrics
References
Anker A, Komai T, Marin IN (2015). A new echiuran associated snapping shrimp (Crustacea: Decapoda: Alpheidae) from the Indo-West Pacific. Zootaxa 3914:441-455. https://doi.org/10.11646/zootaxa.3914.4.4
Arnold K, Bordoli L, Kopp J, Schwede T (2006). The SWISS-Ma web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. https://doi.org/10.1093/bioinformatics/bti770
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37(2):202-208. https://doi.org/10.1093/nar/gkp335
Benkert P, Biasini M, Schwede T (2011). Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343-350. https://doi.org/10.1093/bioinformatics/btq662
Berry EA, Guergova-Kuras M, Huang LS, Crofts AR (2000). Structure and function of cytochrome bc complexes. Annual Review of Biochemistry 69:1005-1075. https://doi.org/10.1146/annurev.biochem.69.1.1005
Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T (2009). Protein structure homology modelling using SWISS-MODEL workspace. Nature Protocols 4:1-13. https://doi.org/10.1038/nprot.2008.197
Chace Jr. FA (1988). The caridean shrimps (Crustacea: Decapoda) of the Albatross Philippine Expedition, 1907-1910, Part 5: Family Alpheidae. Smithsonian Contributions to Zoology 466:1-99.
Chen QL, Tang XS, Yao WJ, Lu SQ (2009). Bioinformatics analysis the complete sequences of cytochrome b of Takydromus sylvaticus and modeling the tertiary structure of encoded protein. International Journal of Biological Sciences 5(6):596-602. https://doi.org/10.7150/ijbs.5.596
Ebenezer NS, Uma Maheswari M, Joyce Priyakumari C (2005). Homology modeling of cytochrome b from Carangoides equula. International Journal of Science and Research 6(2):92-95.
Esposti MD, De Vries S, Crimi M, Ghelli A, Patarnello T, Meyer A (1993). Mitochondrial cytochrome b: evolution and structure of the protein. Biochimica et Biophysica Acta 1143(3):243-271. https://doi.org/10.1016/0005-2728(93)90197
Gao XG, Wen XL, Esser L, Quinn B, Yu L, Yu CA, Xia D (2003). Structural basis for the quinone reduction in the bc(1) complex: A comparative analysis of crystal structures of mitochondrial cytochrome bc(1) with bound substrate and inhibitors at the Qi site. Biochemistry 42(30):9067-9080. https://doi.org/10.1021/bi0341814
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005). Protein identification and analysis tools on ExPASy server. In: Walker JM (Ed). Proteomics Protocol Handbook. Humana Press, pp 571-607.
Geourjon C, Deléage G (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11:681-684. https://doi.org/10.1038/s41598-017-07064-4
Ghosh R, Upadhyay AD, Roy AK, Tiwari A (2020). Structural and functional analysis of cytochrome b protein of Indian major carps Labeo rohita. Journal of Entomology and Zoology Studies 8(2):540-550.
Guex N, Peitsch MC (1997). SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18(15):2714-2723. https://doi.org/10.1002/elps.1150181505
Gupta A, Pal SK, Narayan J, Potukuchi SM (2009). In silico structure prediction of p27SJ, a novel protein in St John’s Wort, that suppresses expression of HIV-1 genome. Biofrontiers 1:32-35.
Hacker B, Barquera B, Crofts AR, Gennis RB (1993). Characterization of mutations in the cytochrome-b subunit of the bc(1)-complex of Rhodobacter sphaeroides that affect the quinone reductase site (Qc). Biochemistry 32(16):4403-4410. https://doi.org/10.1021/bi00067a033
Idicula-Thomas S, Balaji PV (2005). Understanding the relationship between the primary structure of proteins and their amyloidogenic propensity: clues from inclusion body formation. Protein Engineering, Design and Selection 18:175-180. https://doi.org/10.1093/protein/gzi022
Jha PN, Purushothaman P, Chinnadurai S, Renjith RK, Remesan MP, Baiju MV, Edwin L (2019). Occurrence of Alpheus euphrosyne de Man, 1897 (Crustacea: Decapoda: Alpheidae) from the South-Eastern Arabian Sea, India. Indian Journal of Geo Marine Sciences 48(11):1695-1698.
Kyte J, Doolittle RF (1982). A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology 157:105-142.
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996). AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. Journal of Biomolecular NMR 8:477-486. https://doi.org/10.1007/ bf00228148
Lovell SC, Davis IW, Arendall WB, Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC (2003). Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins: Structure Function and Bioinformatics 50(3):437-450
Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Sali A (2000). Comparative protein structure modeling of genes and genomes. Annual Review of Biophysics and Biomolecular Structure 29:291-325. https://doi.org/10.1146/annurev.biophys.29.1.291
Schrodinger L, DeLano W (2020). PyMol. Retrieved from http://www.pymol.org/pymol
Singh M, Gupta A, Lakra WS (2012). In silico 3-D structure prediction of cytochrome b protein of sisorid catfish Glyptothorax ngapang. Indian Journal of Biotechnology 11:156-162.
Studer G, Rempfer C, Waterhouse AM, Gumienny G, Haas J, Schwede T (2020). QMEANDisCo - distance constraints applied on model quality estimation. Bioinformatics 36:1765-1771. https://doi.org/10.1093/bioinformatics/btz828
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, … Mering CV (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research 43:D447-D452.
Tamura K, Dudley J, Nei M, Kumar M (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software v. 4.0. Molecular Biology and Evolution 24:1596-1599. https://doi.org/10.1093/molbev/msm092
Yan BX, Sun YQ (1997). Glycine residues provide flexibility for enzyme active sites. Journal of Biological Chemistry 272:3190- 3194. https://doi.org/10.1074/jbc.272.6.3190
Downloads
Published
How to Cite
Issue
Section
License
Papers published in Notulae Scientia Biologicae are Open-Access, distributed under the terms and conditions of the Creative Commons Attribution License.
© Articles by the authors; licensee SMTCT, Cluj-Napoca, Romania. The journal allows the author(s) to hold the copyright/to retain publishing rights without restriction.
License:
Open Access Journal - the journal offers free, immediate, and unrestricted access to peer-reviewed research and scholarly work, due SMTCT supports to increase the visibility, accessibility and reputation of the researchers, regardless of geography and their budgets. Users are allowed to read, download, copy, distribute, print, search, or link to the full texts of the articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author.













.png)















