Computational study of environmental stress-related transcription factor binding sites in the promoter regions of maize auxin response factor (ARF) gene family
DOI:
https://doi.org/10.15835/nsb12310823Keywords:
expression study; heatmap; transcriptional factor binding site; zmARF; Zea maysAbstract
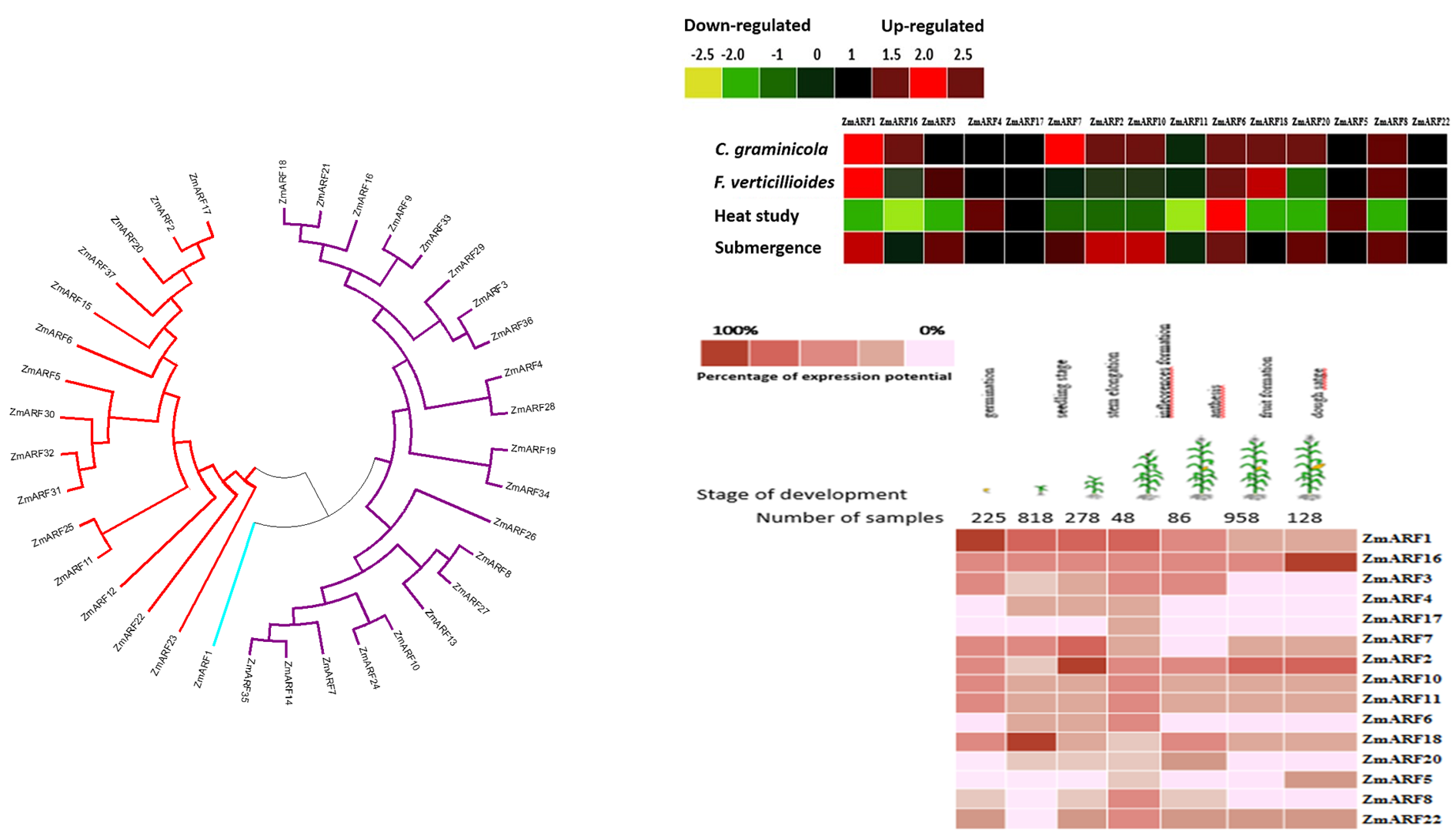
Auxin response factors (ARF) gene family plays key roles in plant development and act as transcription factors (TFs) in the regulation of gene expression. An extensive bioinformatics analysis including analysis of conserved motifs, chromosomal map, phylogenetic relationships, and expression profiles were performed for the maize ARF gene family. In this study, a set of publicly available 38 ARF maize (Zea mays) nucleotide sequences were downloaded. Using microarray data, a bioinformatics search for identification of TFBs in ARF genes using plant promoter analysis (PlantPAN) was carried out. The 38 maize ARF genes were categorized into three groups (Class I, II, and III). ARF genes have been studied by molecular methods in several different plant species however to better understand the mechanisms of these genes more studies are needed. Gene cluster analysis showed that the same set of genes on the chromosomes were positively correlated with the same number of gene clusters. Several TFBs including AP2/ERF, ERF, WRKY, bZIP, bHLH, GATA, and NAC were identified in the promoter regions. These TFBs are responsible for modulation of several biotic stress-responsive genes. The main aims of the present study were to obtain genomic information for the ZmARF gene family and their expression under abiotic and biotic stresses.
Metrics
References
Berrocal-Lobo M, Molina A, Solano R (2002). Constitutive expression of Ethylene-response-factor1 in Arabidopsis confers resistance to several necrotrophic fungi. The Plant Journal 29:23-32. https://doi.org/10.1046/j.1365-313x.2002.01191.x
Blanco E, Messeguer X, Smith TF, Guigó R (2006). Transcription factor map alignment of promoter regions. PLoS Computational Biology 2(5):e49. https://doi.org/10.1371/journal.pcbi.0020049
Bouzroud S, Gouiaa S, Hu N, Bernadac A, Mila I, Bendaou N, … Zouine M (2018). Auxin response factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PloS One 13(2): e0193517. https://doi.org/10.1371/journal.pone.0193517
Chu X, Wang C, Chen X, Lu W, Li H, Wang X, … Guo X (2015). The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS One 10(11):e0143022. https://doi.org/10.1371/journal.pone.0143022
de Jesus Miranda V, Porto WF, da Rocha Fernandes G, Pogue R, Nolasco DO, Araujo AC, … Franco OL (2017). Comparative transcriptomic analysis indicates genes associated with local and systemic resistance to Colletotrichum graminicola in maize. Scientific Reports 7(1):2483. https://doi.org/10.1038/s41598-017-02298-8
Di DW, Zhang C, Luo P, An CW, Guo GQ (2016). The biosynthesis of auxin: how many paths truly lead to IAA? Plant Growth Regulation 78(3):275-85. https://doi.org/10.1007/s10725-015-0103-5
Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW (2005). Auxin response factor1 and auxin response factor2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132:4563-4574. https://doi: 10.1242/dev.02012
Finet C, Fourquin C, Vinauger M, Berne‐Dedieu A, Chambrier P, Paindavoine S, Scutt CP (2010). Parallel structural evolution of auxin response factors in the angiosperms. The Plant Journal 63(6):952-9. https://doi.org/10.1186/s13007-020-00609-1
Fujita Y, Nakashima K, Yoshida T, Fujita M, Shinozaki K, Yamaguchi‐Shinozaki K (2013). Role of abscisic acid signaling in drought tolerance and preharvest sprouting under climate change. Climate Change and Plant Abiotic Stress Tolerance 18:521-54. https://doi.org/10.1002/9783527675265.ch20
Goetz M, Hooper LC, Johnson SD, Rodrigues JC, Vivian-Smith A, Koltunow AM (2007). Expression of aberrant forms of auxin response factor8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiology 145(2):351-66. https://doi.org/10.1111/j.1365-313X.2010.04292.x
Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM (2006). Auxin response factor8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18(8):1873-86. https://doi.org/10.1105/tpc.105.037192
Greenham K, McClung CR (2015). Integrating circadian dynamics with physiological 649 processes in plants. Nature Reviews Genetics 16:598-610. https://doi.org/10.1038/nrg3976
Guilfoyle TJ, Hagen G (2012). Getting a grasp on domain III/IV responsible for Auxin Response Factor–IAA protein interactions. Plant Science 190:82-88. https://doi.org/10.1016/j.plantsci.2012.04.003
Hagen G, Guilfoyle T (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Molecular Biology 49(3-4):373-85. https://doi.org/10.1023/A:1015207114117
Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, … Goda H (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proceedings of the National Academy of Sciences 104(15):6478-83. https://doi.org/10.1073/pnas.0611629104
Hogeweg P (2011). The roots of bioinformatics in theoretical biology. PLoS Computational Biology 7(3):e1002021. https://doi.org/10.1371/journal.pcbi.1002021
Hunter C, Willmann, MR, Wu G, Yoshikawa M, de la Luz Gutiérrez-Nava M, Poethig SR (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133(15):2973-81. https://doi.org/10.1242/dev.02491
Ito J, Fukaki H, Onoda M, Li L, Li C, Tasaka M, Furutani M (2016). Auxin-dependent compositional change in Mediator in ARF7-and ARF19-mediated transcription. Proceedings of the National Academy of Sciences 113(23):6562-5657. http://doi.org/10.1073/pnas.1600739113
Jiang W, Wu J, Zhang Y, Yin L, Lu J (2015). Isolation of a WRKY30 gene from Muscadinia rotundifolia (Michx) and validation of its function under biotic and abiotic stresses. Protoplasma 252(5):1361-74. https://doi.org/10.1007/s00709-015-0769-6
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology 17(3):287. https://doi.org/10.1038/7036
Khurana JP, Tamot BK, Maheshwari SC (1988). Floral Induction in a photo periodically insensitive duckweed, Lemna paucicostata LP6: role of glutamate, aspartate, and other amino acids and amides. Plant Physiology 86(3):904-907. https://doi.org/10.1104/pp.86.3.904
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, … Rombauts S (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30(1):325-257. https://doi.org/10.1093/nar/30.1.325
Li SB, Ou Yang WZ, Hou XJ, Xie LL, Hu CG, Zhang JZ (2015). Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Frontiers Plant Science 30(6):119. https://doi.org/10.3389/fpls.2015.00119
Liao Y, Liu S, Jiang Y, Hu C, Zhang X, Cao X, … Chen R (2017). Genome-wide analysis and environmental response profiling of dirigent family genes in rice (Oryza sativa). Genes Genomics 39(1):47-62. https://doi.org/10.1007/s13258-016-0474-7
Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, Woo HR, Nam HG (2010). Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. Journal of Experimental Botany 61(5):1419-30. https://doi.org/10.1093/jxb/erq010
Liu Y, Jiang H, Chen W, Qian Y, Ma Q, Cheng B, Zhu S (2011). Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regulation 63(3):225-234. https://doi.org/10.1007/s10725-010-9519-0
Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, … Ecker JR (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132(18):4107-4118. http://doi.org/10.1242/dev.01955
Rahaie M, Gomarian M, Alizadeh H, Malboobi MA, Naghavi MR (2011). The expression analysis of transcription factors under long term salt stress in tolerant and susceptible wheat genotypes using reverse northern blot technique. Iranian Journal of Crop Sciences 13(3):580-595.
Ren Z, Liu R, Gu W, Dong X (2017). The Solanum lycopersicum auxin response factor SlARF2 participates in regulating lateral root formation and flower organ senescence. Plant Science 256:103-111. http://doi.org/10.3389/fpls.2018.01395
Saidi A, Hajibarat Z (2018). In silico analysis of floral MADS-BOX gene in Brachypodium distachyon. Bionature 366-375.
Saidi A, Hajibarat Z (2019). Characterization of cis-elements in hormonal stress-responsive genes in Oryza sativa. Asia-Pacific Journal Molecular Biology and Biotechnology 27(1):95-102. https://doi:10.35118/apjmbb.2019.027.1.10
Saidi A, Hajibarat Z, Hajibarat Z (2020). Transcriptome analysis of Phytophthora infestans and Colletotrichum coccodes in tomato to reveal resistance mechanisms. Asia-Pacific Journal Molecular Biology and Biotechnology. https://doi.org/10.35118/apjmbb.2020.028.1.05
Sun R, Wang K, Guo T, Jones DC, Cobb J, Zhang B, Wang Q (2015). Genome-wide identification of auxin response factor (ARF) genes and its tissue-specific prominent expression in Gossypium raimondii. Functional & Integrative Genomics 15(4):481-93. https://doi.org/10.1007/s10142-015-0437-0
Tabata R, Ikezaki M, Fujibe T, Aida M, Tian CE, Ueno Y, … Ishiguro S (2010). Arabidopsis auxin response factor 6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant and Cell Physiology 51(1):164-175.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30(12):2725-2729.https://doi.org/10.1093/molbev/mst197
Tiwari SB, Hagen G, Guilfoyle T (2003). The roles of auxin response factor domains in auxin-responsive transcription. The Plant Cell 15(2):533-43. https://doi.org/10.1105/tpc.008417
Voorrips RE (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93(1):77-8. https://doi.org/10.1093/jhered/93.1.77
Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY (2005). Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. The Plant Cell 7(8):2204-2216. https://doi.org/10.1105/tpc.105.033076
Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signalling pathway. Current Biology 17:1784-1790. https://doi.org/10.1016/j.cub.2007.09.025
Wang R, Estelle M (2014). Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Current Opinion in Plant Biology 21:51–58. https://doi.org/10.1016/j.pbi.2014.06.006
Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, … Reed JW (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin‐induced lateral root formation. The Plant Journal 43(1):118-30. https://doi.org/10.1111/j.1365-313X.2005.02432.x
Yang Y, Zhou Y, Chi Y, Fan B, Chen Z (2017). Characterization of soybean WRKY gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode. Scientific Reports 7(1):17804. https://doi.org/10.1038/s41598-017-18235-8
Zheng Z, Qamar SA, Chen Z, Mengiste T (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. The Plant Journal 48(4):592-605. https://doi.org/10.1111/j.1365-313X.2006.02901.x
Zouine M, Fu Y, Chateigner-Boutin AL, Mila I, Frasse P, Wang H, … Bouzayen M (2014). Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PloS One 9(1):e84203. https://doi.org/10.1371/journal.pone.0084203
Downloads
Published
How to Cite
Issue
Section
License
Papers published in Notulae Scientia Biologicae are Open-Access, distributed under the terms and conditions of the Creative Commons Attribution License.
© Articles by the authors; licensee SMTCT, Cluj-Napoca, Romania. The journal allows the author(s) to hold the copyright/to retain publishing rights without restriction.
License:
Open Access Journal - the journal offers free, immediate, and unrestricted access to peer-reviewed research and scholarly work, due SMTCT supports to increase the visibility, accessibility and reputation of the researchers, regardless of geography and their budgets. Users are allowed to read, download, copy, distribute, print, search, or link to the full texts of the articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author.













.png)















