Molecular variation and phylogenetic status of ponyfish (Perciformes: Leiognathidae) in Karaikal, South India
DOI:
https://doi.org/10.15835/nsb12210661Keywords:
COI; genetic variation; Leiognathidae; NJ tree; nucleotide diversityAbstract
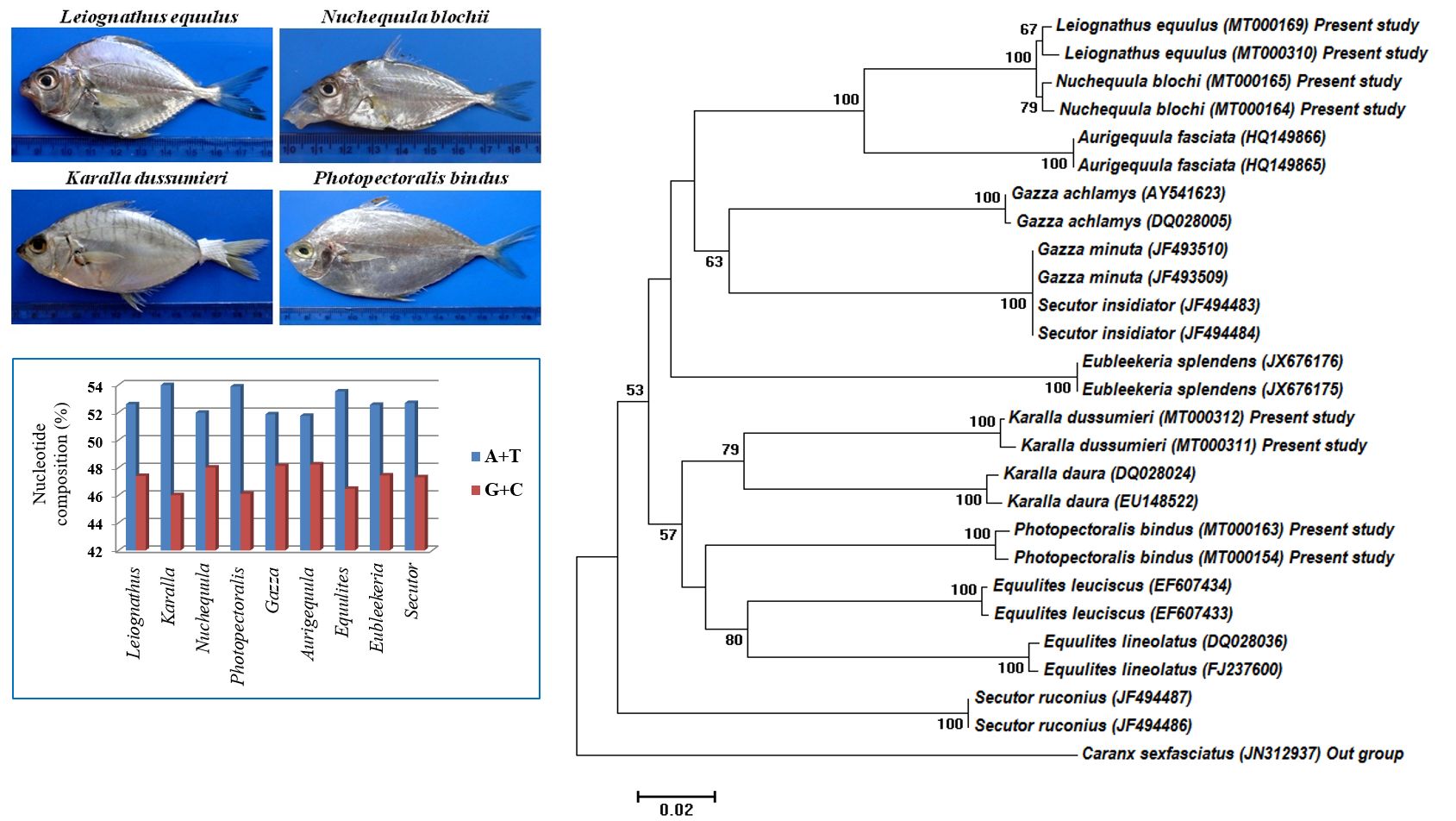
The family Leiognathidae, commonly known as ponyfish and they are mainly classified by mouth morphology. In this study, genetic variation and phylogenetic relationships among nine genera and 13 species of Leiognathidae, inferred from Cytochrome Oxydase I gene. The Neighbour Joining tree showed the paraphyly of the studied species form genus Secutor and monophyly status all the other eight genera. Leiognathus equulus is occupying a basal branch of the family NJ tree. The highest genetic distance (0.349) showed between Eubleekeria and Nuchequula. The lowest genetic distance (0.007) showed between the genus Nuchequula and Leiognathus. Among the nine genera, very less nucleotide diversity (π= 0.1100) was in Secutor and higher value (π = 0.7558) was recorded in Leiognathus. The Tajima’s test statistic (D) value was positive, so, the entire genus is in balancing selection.
Metrics
References
Abraham KJ, Joshi KK, Murty VSR (2011). Taxonomy of the fishes of the family Leiognathidae (Pisces, Teleostei) from the West coast of India. Zootaxa 2886:1-18. https://doi.org/10.11646/zootaxa.2886.1.1
Chakrabarty P, Sparks JS (2008). Diagnoses for Leiognathus Lacepede 1802, Equula Cuvier 1815, Equulites Fowler 1904, Eubleekeria Fowler 1904, and a new ponyfish genus (Teleostei: Leiognthidae). American Museum Novitates 3623:1-11. https://doi.org/10.1206/618.1
Chakrabarty P, Amarasinghe T, Sparks JS (2008). Redescription of ponyfishes (Teleostei: Leiognathidae) of Srilanka and the status of Aurigequula Fowler 1918. Ceylon Journal of Science (Biological Sciences) 37(2):143-161. https://doi.org/10.4038/cjsbs.v37i2.502
Costa FO, Carvalho GR (2007). The barcode of life initiative: synopsis and prospective societal impacts of DNA barcoding of fish. Genomics Society and Policy 3:29-40. https://doi.org/10.1186/1746-5354-3-2-29
Eschmeyer WN (2010). Catalog of fishes. Electronic version (Updated 25th October 2010). http://research.calacademy.org/research/ichthyology/catalog/fishcatsearch.html
Froese R, Pauly D (2017). Fish Base. World Wide Web Electronic Publication. www.fishbase.org
Hebert PDN, Cywinska A, Ball SL, Waard JRD (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society of London. Series B: Biological Sciences 270:313-321. https://doi.org/10.1098/rspb.2002.2218
Ikejima K, Ishiguro NB, Wada M, Tsukamoto K, Nishida M (2004). Molecular phylogeny and possible scenario of ponyfish (Perciformes: Leiognathidae) evolution. Molecular Phylogenetic and Evolution 30:904-909. https://doi.org/10.1016/j.ympev.2003.10.006
Kaeding AJ, Ast JC, Pearce MM, Urbanczyk H, Kimura S, Endo H, … Dunlap PV (2007). Phylogenetic diversity and cosymbiosis in the bioluminescent symbioses of Photobacterium mandapamensis. Applied and Environmental Microbiology 73:3173-3182. https://doi.org/10.1128/aem.02212-06
Lakra WS, Verma MS, Goswami M, Lal KK, Mohindra V, Punia P, … Hebert P (2011). DNA barcoding Indian marine fishes. Molecular Ecology Resources 11:60-71. https://doi.org/10.1111/j.1755-0998.2010.02894.x
Lynch M, Crease TJ (1990). The analysis of population survey data on DNA sequence variation. Molecular Biology and Evolution 7(4):377-394. https://doi.org/10.1093/oxfordjournals.molbev.a040607
Peris M, Chandra Sekhar Reddy A, Rao LM, Khedkar GD, Ravinder K, Nasruddin K (2009). COI (Cytochrome oxi¬dase-I) sequence-based studies of Carangid fishes from Kakinada coast, India. Molecular Biology Reports 36:1733-1740. https://doi.org/10.1007/s11033-008-9375-4
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003). Dna SP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19(18):2496-2497. https://doi.org/10.1093/bioinformatics/btg359
Saitou N, Nei M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4(4):406-425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sambrook J, Fritsch EF, Maniatis T (1989). Molecular cloning: a laboratory manual, second ed. Cold Spring Harbor Laboratory Press, New York, Cold Springs Harbor, pp 1626.
Seah YG, Ghaffar MA, Usup G (2008). Phylogeny of ponyfishes from coastal waters of the South China Sea. Journal of Applied Biological Sciences 2(3):125-133.
Sparks JS, Dunlap PV (2004). A clade of non-sexually dimorphic ponyfishes (Teleostei: Perciformes: Leiognathidae): phylogeny, taxonomy and description of a new species. American Museum Novitates 3459:1-21. https://doi.org/10.1206/0003-0082(2004)459<0001:acondp>2.0.co;2
Sparks JS, Dunlap PV, Smith WL (2005). Evolution and diversification of a sexually dimorphic luminescent system in ponyfishes (Teleostei: Leiognathidae), including diagnoses including diagnoses for two new genera. Cladistics 21:305-327. https://doi.org/10.1111/j.1096-0031.2005.00067.x
Tajima F (1989). Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 123:585-595. https://pubmed.ncbi.nlm.nih.gov/2513255/
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28:2731-2739. https://doi.org/10.1093/molbev/msr121
Ward RD, Zemlac TC, Innes BH, Last PR, Hebert PDN (2005). DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B: Biological Sciences 360(1462):1847-1857. https://doi.org/10.1098/rstb.2005.1716
Downloads
Published
How to Cite
Issue
Section
License
Papers published in Notulae Scientia Biologicae are Open-Access, distributed under the terms and conditions of the Creative Commons Attribution License.
© Articles by the authors; licensee SMTCT, Cluj-Napoca, Romania. The journal allows the author(s) to hold the copyright/to retain publishing rights without restriction.
License:
Open Access Journal - the journal offers free, immediate, and unrestricted access to peer-reviewed research and scholarly work, due SMTCT supports to increase the visibility, accessibility and reputation of the researchers, regardless of geography and their budgets. Users are allowed to read, download, copy, distribute, print, search, or link to the full texts of the articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author.













.png)















