Phylogenetic Analysis and In Silico Characterization of Cytochrome P450 1A (Cyp1A) Protein from the African Catfish, Clarias gariepinus (Burchell, 1822)
DOI:
https://doi.org/10.15835/nsb11410475Keywords:
catfish; Clarias gariepinus; CYP1A; phylogeny; Ramachandran plotAbstract
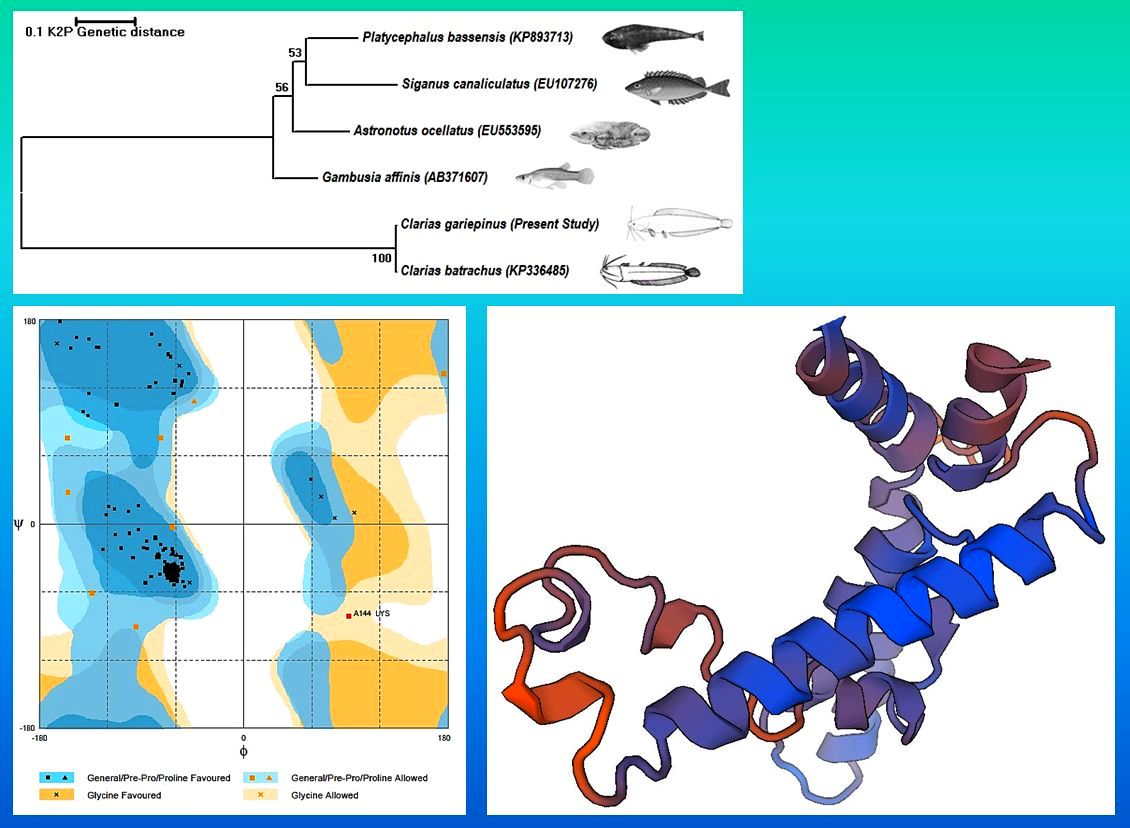
The CYP family enzymes are broadly used as biomarkers because of their pattern of expression. This study describes the application of in silico tools to predict the physico-chemical characters of CYP1A protein from the catfish, Clarias gariepinus. The nucleotide sequence analysis of C. gariepinus CYP1A gene showed higher similarity with C. batrachus and reflected in the phylogenetic tree. The comparative modelling results showed this CYP1A protein was highly similar with the 3-D crystal structure of human Cytochrome p450 1A1 (PDB: 1BE3). The prediction results depicted that most of the amino acids formed alpha helix. The predicted pI was 9.10, hydropathycity was -0.226, exposed and buried residues were 61.67, 38.33% respectively. Ramachandran plot analysis showed that most of the amino acids falling on the favoured region and exhibited right- handed alpha helices as the most stable secondary structure. Some amino acids were also found to form loops to interconnect different helices. The CYP1A protein was predicted to be localized in the mitochondrion of the eukaryotic cell.
Metrics
References
Brammell BF, McClain JS, Oris JT, Price DJ, Birge WJ, Elskus AA (2010). CYP1A expression in caged rainbow trout discriminates among sites with various degrees of polychlorinated biphenyl contamination. Archives of Environmental Contamination and Toxicology 58(3):772-782.
de Moor IJ, Bruton MN (1988). Atlas of alien and translocated indigenous aquatic animals in southern Africa. National Scientific Programmes Unit: CSIR. Report No. 144. Port Elizabeth, South Africa pp 310.
Edwards RJ, Singleton AM, Murray BP, Murray S, Boobis AR, Davies DS (1991). Identification of a functionally conserved surface region of rat cytochromes P4501A. Journal of Biochemistry 278:249-757.
Goksoyr A (1995). Use of cytochrome P450 1A (CYP1A) in fish as a biomarker of aquatic pollution. Archives of Toxicology 17:80-95.
Jayaram KC (1966). Contributions to the study of the fishes of the family Bragidae. Zoological Survey of India 207:1-136.
Jung JH, Kim M, Yim UH, Ha SY, An JG, Won JH, ... Shim WJ (2011). Biomarker responses in pelagic and benthic fish over 1 year following the Hebei Spirit oil spill (Taean, Korea). Marine Pollution Bulletin 62(8):1859-1866.
Kimura M (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16:111-120.
Marti-Renom MA, Stuart AS, Fiser A, Sanchez R, Melo F (2000). Comparative protein structure modelling of genes and genome. Annual Review of Biophysical and Biomolecular Structure 29:291-325.
Nelson JS (1994). Fishes of the world. 3rd Ed. John Wiley and sons. New York pp 622.
Nilsen BM, Berg K, Goksoyr A (1998). Induction of cytochrome P450 1A (CYP1A) in fish. A biomarker for environmental pollution. Methods of Molecular Biology 107:423-438.
Sambrook J, Fritsch EF, Maniatis T (1989). Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, New York.
Sayle R, Milner-White EJ (1995). RasMol: Biomolecular graphics for all. Trends in Biochemical Sciences 20(9):374-376.
Stegeman JJ (1989). Cytochrome P450 forms in fish: Catalytic, immunological and sequence similarities. Xenobiotica 19(10):1093-1110.
Stegeman JJ, Hahn ME (1994). Biochemistry and molecular biology of monooxygenases- current directions in forms, functions, and regulation of cytochrome P450 in aquatic species. In: Malins DC, Ostrander GK (Eds). Aquatic toxicology cellular molecular and biochemical perspectives. CRC Press, Boca Raton, Florida pp 87-206.
Tamura K, Dudley J, Nei M, Kumar M (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software v. 4.0. Molecular Biology and Evolution 24:1596-599.
Williamson AR (2000). Creating a structural genomic consortium. Nature Structural Biology 7(11):953.
Downloads
Published
How to Cite
Issue
Section
License
Papers published in Notulae Scientia Biologicae are Open-Access, distributed under the terms and conditions of the Creative Commons Attribution License.
© Articles by the authors; licensee SMTCT, Cluj-Napoca, Romania. The journal allows the author(s) to hold the copyright/to retain publishing rights without restriction.
License:
Open Access Journal - the journal offers free, immediate, and unrestricted access to peer-reviewed research and scholarly work, due SMTCT supports to increase the visibility, accessibility and reputation of the researchers, regardless of geography and their budgets. Users are allowed to read, download, copy, distribute, print, search, or link to the full texts of the articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author.













.png)















